Current Research Projects
Below you’ll find a list of research projects our team are currently working on. You may as well read about our past projects.
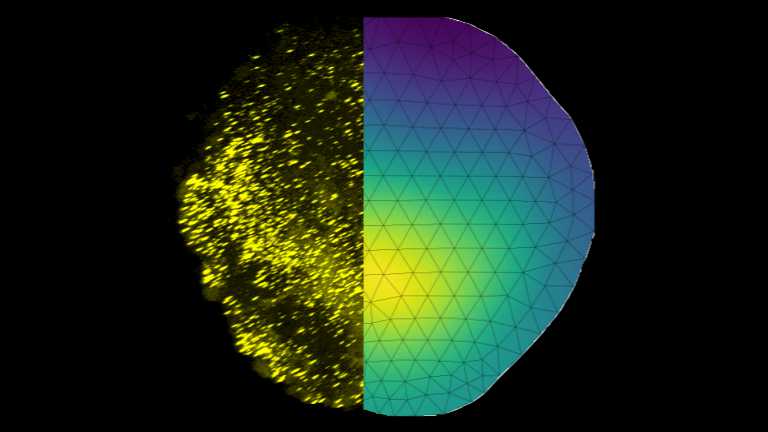
Physical principles of pattern scaling during fin regeneration in zebrafish
Maximilian Kotz
The zebrafish is known for its ability to regenerate lost body parts, including fins. The pectoral fin of larval zebrafish is an ideal model system for studying regeneration because it can be easily manipulated, is amenable to high-resolution imaging and its regeneration is fast. We are close collaborating with the Rita Mateus lab, (Physics of Life, TU Dresden) to understand principles of regeneration, growth, and size control.
Our theoretical research aims to develop mathematical models to understand the role of BMP morphogen gradients regulating fin growth. The ability of gradients to scale with the size of the tissue seems to be one key aspect of robust growth. Within two days, the size of the fin can double, with final size controlled precise up to a few percent.
The methods we use include analyzing fluorescence images, building mathematical models, e.g., reaction-diffusion models in several dimensions, and iteratively refining these models guided by data. Ultimately, we aim to understand physical principles guiding regeneration.
Mathematical modeling of axolotl limb regeneration
Natalia Lyubaykina
Salamanders possess remarkable abilities to regenerate fully functional body parts and organs, which is still beyond modern regenerative medicine capabilities. Thus, they are important model organisms to study regeneration.
My project focuses on limb regeneration process in axolotl salamanders. While key signaling factors controling limb regeneration are known (e.g., morphogens). we lack mechanistic understanding how morphogen gradients form and control growth, let alone how limbs of proportional size regenerate in different-sized animals. Our experimental collaborators Elly Tanaka (IMP Vienna) and Tatiana Sandoval-Guzman (TU Dresden) use quantitative imaging of morphogen dynamics, which guides my mathematical modeling.
Specifcally, my research comprises modelling of reaction-diffusion kinetics coupled to local tissue growth and development of image analysis algorithms to quantify the spatio-temporal patterns of morphogens dynamics, thus constraining and parametrizing the mathematical models.
Cellular navigation
Julian Rode
Biological system process information despite noise-corrupted input, often operating at limits imposed by physics. A prime example is chemotaxis, i.e., active navigation in spatial fields of chemical cues, which enables immune cells to find inflammation sites, sperm cells to find the egg, and bacteria and social amoeba to build communities. Intriguingly, cells of different size use different chemotaxis strategies, comparing concentrations in either space or time. Only heuristic arguments exist to explain this evolutionary choice. We developed an information theory of an ideal agent that combines both strategies allows to quantify 'chemotaxis in bits', to predict when each strategy provides more information.
Read more:

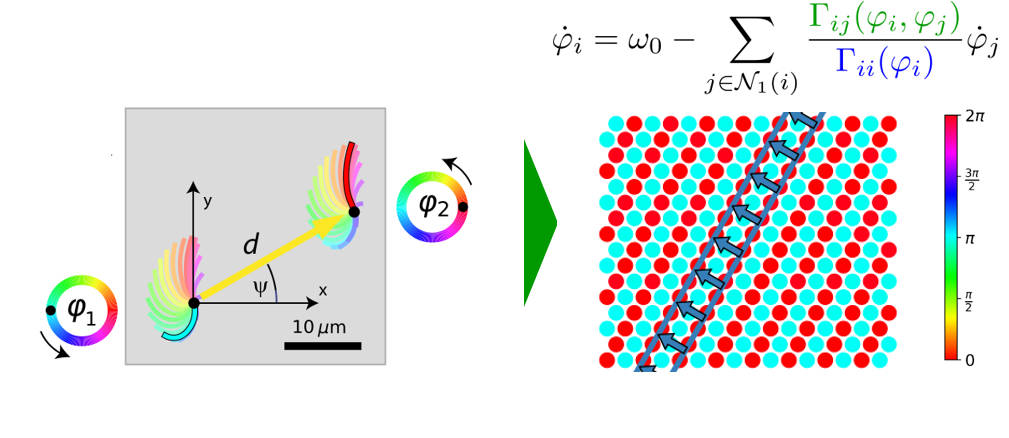
Cilia synchronization
Anton Solovev
Motile cilia are slender cell appendages found on the surface of mammalian airways, brain ventricles, and the oviduct. These cilia beat rhythmically, often in a coordinated fashion, to pump fluids such as mucus or cerebrospinal fluid effectively. This metachronal coordination in cilia carpets , similar to a Mexican wave in a soccer stadium, presumably originate from hydrodynamic interactions between neighboring cilia. Yet, basic questions like “how does the direction of metachronal waves is set by the shape of the cilia beat” are open.
We study metachronal coordination in cilia carpets from a physicists' perspective, modeling each cilium as a noisy phase oscillator, coupled to its neighbors (similar to the Kuramoto model with local coupling). Using multi-scale simulations and theory, we answer questions of multi-stability of metachronal waves, as well as their robustness to active fluctuations.
Read more
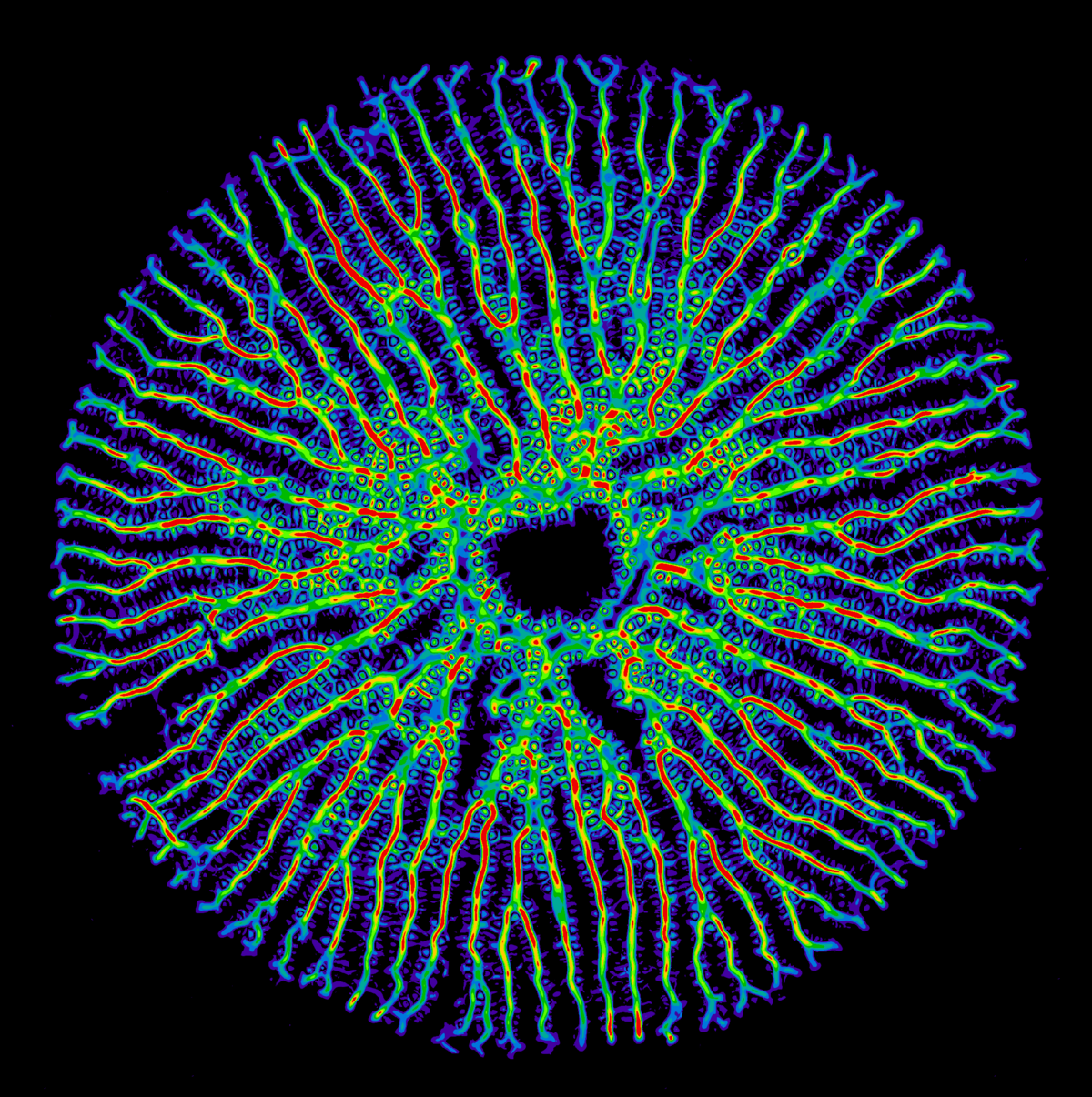
Valve pattern formation in diatoms
Iaroslav Babenko
Diatoms are a large group of single-celled algae that are prominent for their ability to deposit silicic acid into intricate patterns of biosilicified cell walls: diatoms live in glass houses. Their cell walls display a bewildering architectural complexity with hierarchical pore patterns, ribs and spikes. Yet, the physical mechanisms that shape these lightweight structures remain poorly understood. Recent computational models can explain biosilification at the micro-scale, but not yet the emergence of regular patterns on the meso-scale.
My project combines theory and experiment to understand the mechanism of valve pattern formation, using Thalassiosira pseudonana as a model organism. Currently, we investigate an unusual reaction-diffusion model that generates growing rib patterns as observed experimentally inside the silica deposition vesicle of dividing diatoms, where new valves form de novo. In parallel, we perform electron microscopy of developing valves and establish automated image analysis algorithms to characterize the morphodynamics of biosilica valves at different developmental stages.
Our aim is to identify possible mechanisms of self-organized pattern formation at the meso-scale during cell wall biogenesis in diatoms.
Read more:
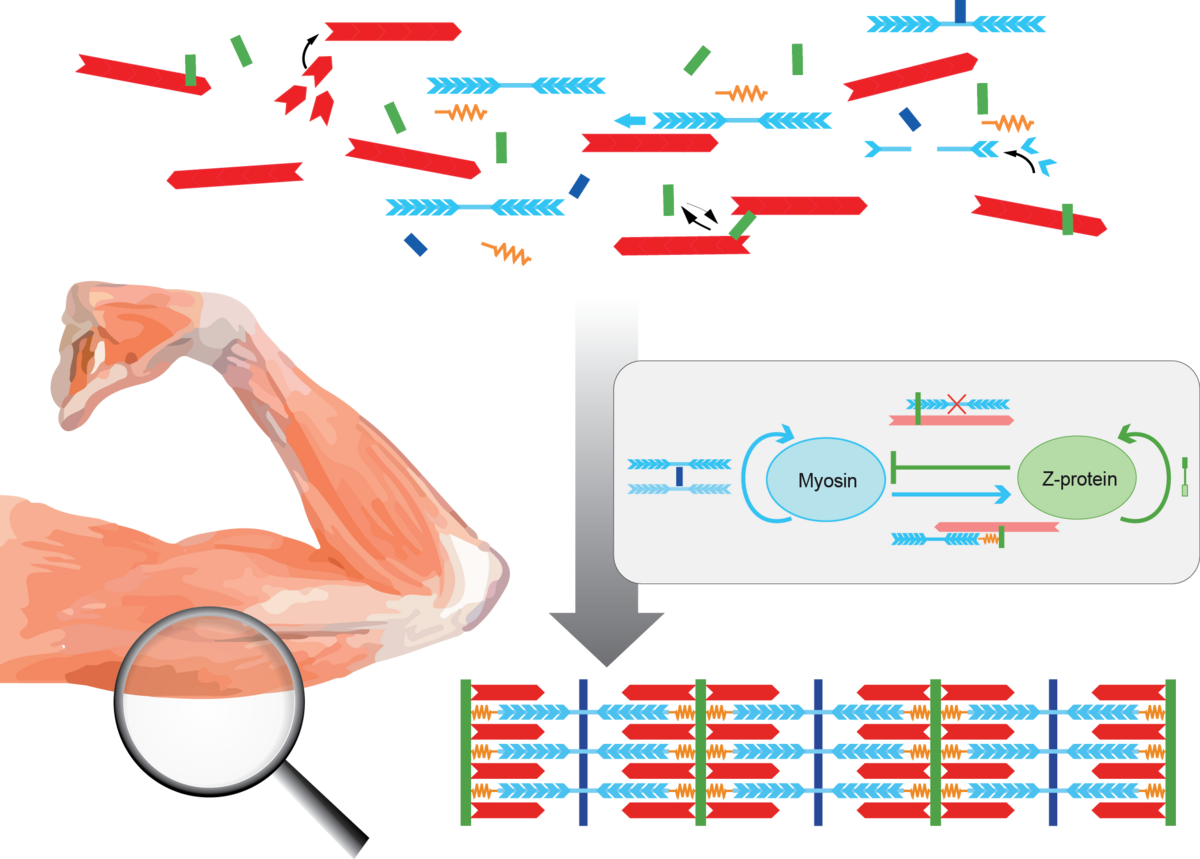
Mathematical modeling of developing muscles
Francine Kolley
How to build a muscle? Striated muscles are organized in pseudo-crystalline structures, called sarcomeres. They contain three different types of filaments. Sarcomeres consist of polar actin filaments crosslinked at their structural plus ends to the Z-disc, while their minus ends face the center of the sarcomere, where they interact with anchored bipolar myosin molecular motors. The giant protein Titin serves as an elastic spring that links the actin and myosin filaments. We do not understand how these elaborate structures self-assemble during muscle development.
We develop theoretical models of how cytoskeletal filaments in an initial disordered acto-myosin bundle interactions spontaneously form periodic patterns and, eventually, highly regular sarcomeres. Our approach combines mean-field models and agent-based simulations to investigate two putative mechanisms of myofibrillogenesis.
Read more:
Image analysis of developing muscles
Ian Estabrook
We are researching the development of myofibrils during myofibrillogenesis, when the proteins that make up muscles self-organise into sarcomeres, and eventually form in to highly ordered crystal-like structures.
In order to study the formation of myofibrils, we are developing Matlab-based feature detection algorithms. Existing feature detection algorithms work well to track fully developed muscles, however, these typically rely on the very regular final patterns to identify sarcomeres. This means these algorithms fail to detect sarcomeres well during myofibrillogenesis. Instead, we use a more general approach combining information from multiple channels from fluorescence microscopy to identify both individual sarcomeres and larger myofibrils. We apply our algorithms to multi-channel fluorescence images of Drosophila during myofibrillogenesis, provided by experimental collaboration partners from the Schnorrer lab (IBDM, Marseilles).
By tracking large numbers of sarcomeres in these images, we hope to identify key features revealing how muscles change during the fundamental formation stages, and identify the mechanisms used to build muscles.
Past Research Projects
Read about what we’ve worked on in the past