Past Research Projects
Find information about our current research on the main Projects page.
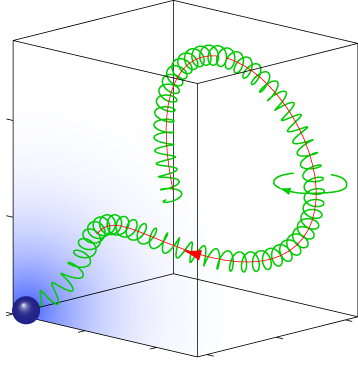
Pathfinding of microswimmers
Purposeful motion requires an orchestrated interplay of motility and sensation. We studied this interplay in the exemplary context of sperm chemotaxis, a process that guides sperm cells on their journey towards the egg. We characterized a navigation strategy along circular and helical paths that is enjoyed by sperm and other microswimmers, but that differs fundamentally from bacterial chemotaxis. This novel navigation strategy is remarkably efficient and robust and reflects perfect adaptation to a noisy environment. Current research aims at understanding quantitativlely the chemotactic signaling system of sperm cells, and to identify general design principles underlying its amazing sensitivity.
Collaboration: Kaupp Lab
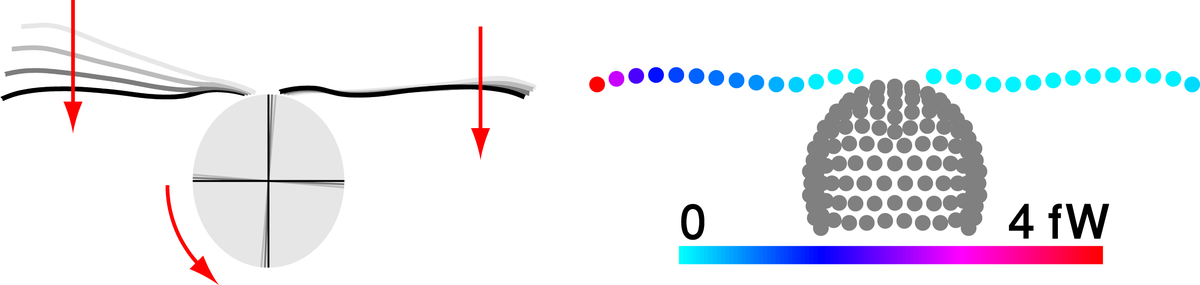
Flagellar synchronization by mechanical control
The eukaryotic flagellum is a best-seller of nature: These slender cell appendages propel sperm and many other microswimmers, including disease-causing protists. In mammalian airways or the oviduct, collections of flagella beat in synchrony to pump fluids efficiently. We use theory and experiment to elucidate a mechanism of synchronization in the model organism Chlamydomonas, a green algal cell that swims with two flagella like a breaststroke swimmer. Our analysis shows how synchronization arises by a coupling of swimming and flagellar beating and characterizes an exemplary force–velocity relationship of the flagellar beat.
Collaboration: Howard Lab
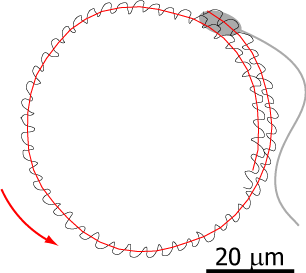
Flagellar propulsion
Flagella are higly conserved cell apdendages that are powered by molecular motors and can beat regularly like a whip. Many microswimmers including sperm cells and algae propel themselves using flagella; in our lungs, epithelial cells use flagella to transport mucus. We study the mechanics of flagellar bending waves and resulting low Reynolds number flows to understand flagellar self-propelsion in two model systems: sperm cells and the flagellated green algae Chlamydomonas. In the past, high-precision measurements of a wiggling motion accompanying sperm swimming allowed us to callibrate its hydrodynamics. From this, we predicted sperm swimming across scales and explained how sperm cells swim in cirlces. This collaboration with the Howard lab at MPI CBG will be continued to study Chlamydomonas swimming.
Collaboration: Howard Lab
Self-organization of robust liver transport networks
The liver represents a “chemical factory” that is characterized by intertwined transport networks for toxins and metabolites. Each hepatocyte cell interacts with two space-filling networks which either transport bile or blood through bile canaliculi or sinusoids. How these networks grow and establish their distinct geometry to supply all cells whilst providing robustness with respect to perturbations remains elusive.
In this project, we elucidate design principles of liver architecture and network self-organization to ultimately understand bile and blood transport in the liver — pursuing a two-fold approach: (i) We analyse high-resolution imaging data of cell membrane compartmentalisation in mouse liver tissue. For this, we propose a nematic tensor formalism to characterise an unconventional type of hepatic cell polarity. Our analysis reveals global patterns of aligned cell polarity on the scale of the liver lobule, the functional unit of the liver, thereby characterizing the three-dimensional liver tissue as a biaxial nematic liquid crystal. We quantify co-alignment of hepatocyte cell polarity and transport network anisotropy. (ii) We then ascertain the functional relevance of this network geometry for robust transport, seeking to elucidate the relationship between local network properties and global transport properties. For that aim, we perform flow simulations in reconstructed bile canaliculi networks as well as in synthetic networks simulated by sets of simple local rules. With this approach that bridges physics and biology, we seek to link structure and function of transport networks in liver tissue.
Collaboration: Zerial Lab
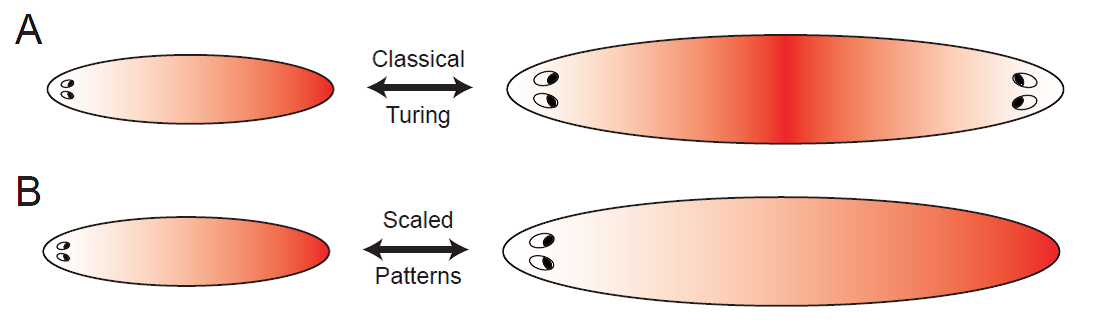
Principles of pattern scaling
During embryonic development and regeneration, multi-cellular organisms face the challenge of robustly patterning their specific body plan adjusted to organism size, using a set of morphogenetic rules. This ability is particularly astonishing in animals capable of regeneration such as flatworms, which can reestablish an appropriately scaled body plan in amputation fragments of arbitrary size. Recently, molecular candidates defining head-tail polarity have been identified.
The simplest model to spontaneously generate head-tail polarity based on graded concentration profiles of signaling molecules is the classical reaction-diffusion system introduced by Turing. However, the resulting patterns do not scale naturally, since diffusion coefficients and reaction rates define fixed characteristic length scales. Together with a talented PhD student, Steffen Werner, we investigate minimal requirements for robust pattern formation systems that are both self-organized, yet, unlike Turing patterns, scale with system size. We propose novel feedback mechanisms that augment classical Turing patterning to allow for robust scaling of reaction-diffusion patterns by self-consistently adjusting its length scale to system size. Thereby, we identify general principles for self-organized and self-scaling pattern formation.
Reference
S Werner, T Stückemann, M Beiran Amigo, JC Rink, F Jülicher, BM Friedrich
Scaling and regeneration of self-organized patterns
Phys. Rev. Lett.114, 2015 – selected as “Editor’s suggestion”
Mechanosensing and the emergence of cytoskeletal order
Tissue cells actively adhere to neighboring cells (or an in vitro substrate) and extert cytoskeletal forces, thus probing their mechanical environment. Substrate stiffness can guide cytoskeletal reorganization, driving the parallel alignment of actin bundles (nematic order) and the formation of acto-myosin crystals (so-called myofibrils) in developing muscle cells. To understand the emergence of cytoskeletal order, we develop minimal descriptions of interacting cytoskeletal force generators. We envision a hierarchy of different actin order that underlies the formation of force-generating myofibrils in muscle cells and thus theoretically address the ultimate question: How to build and regenerate muscle?
Collaboration: Discher Lab