Chromatin at Large Scales
Loop Extrusion and Overall Nuclear Organization

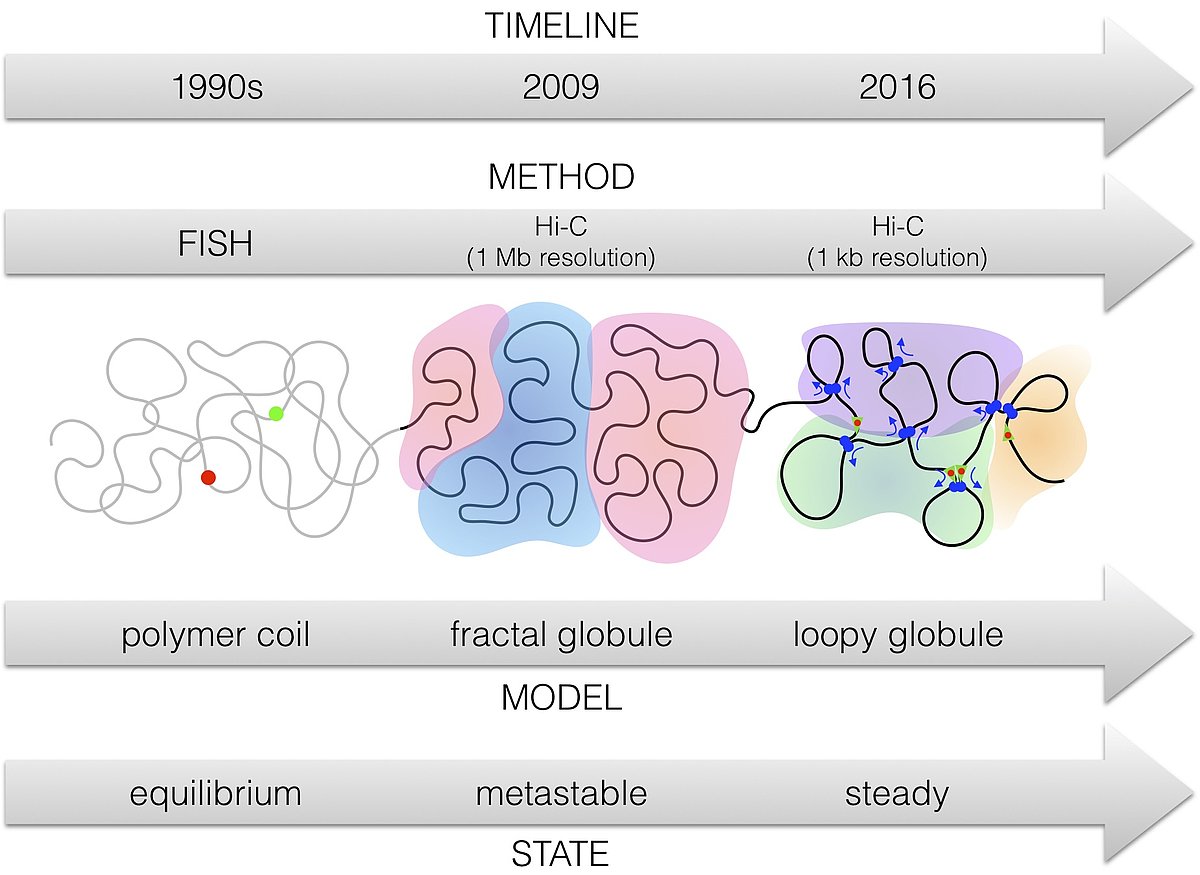
There is a lot of recent progress on the understanding of chromatin at large scales. New experimental methods, based on e.g. chromatin conformation capture, have changed our polymer physics view of chromatin at large scales. Whereas in the 1990s it was an equilibrium view, this has changed in 2009 to a non-equilibrium picture, the fractal globule. In the meantime, chromatin conformation capture at higher resolution has led rather to a steady-state picture, something like a loopy globule, a polymer decorated with loop extruding motor complexes, see (Sazer 2018) for a review.
We address various questions that these new experiments raise, including whether collapsed globules are stable (Schram 2013), whether active processes drive the overall nucleus structure (Yamamoto 2017a), what are possible mechanisms driving loop extrusion (Yamamoto 2017b), what type of polymer dynamics is caused by loop extrusion (Yamamoto 2019a), and how it influences phase-separated liquid droplets of transcriptional activators (Yamamoto 2019b).