The Mechanical Genome
Nucleosome Positioning Rules and Multiplexing

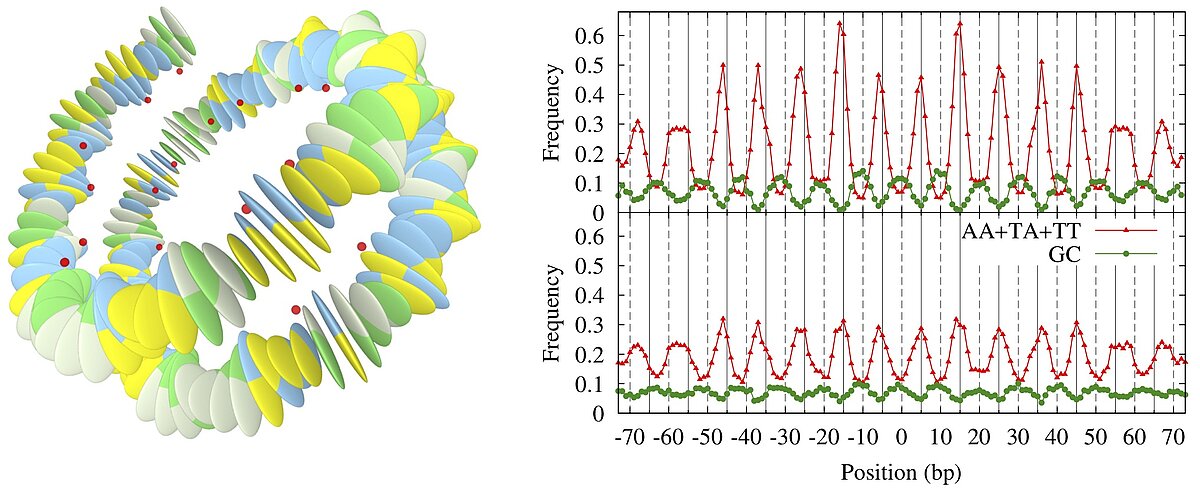
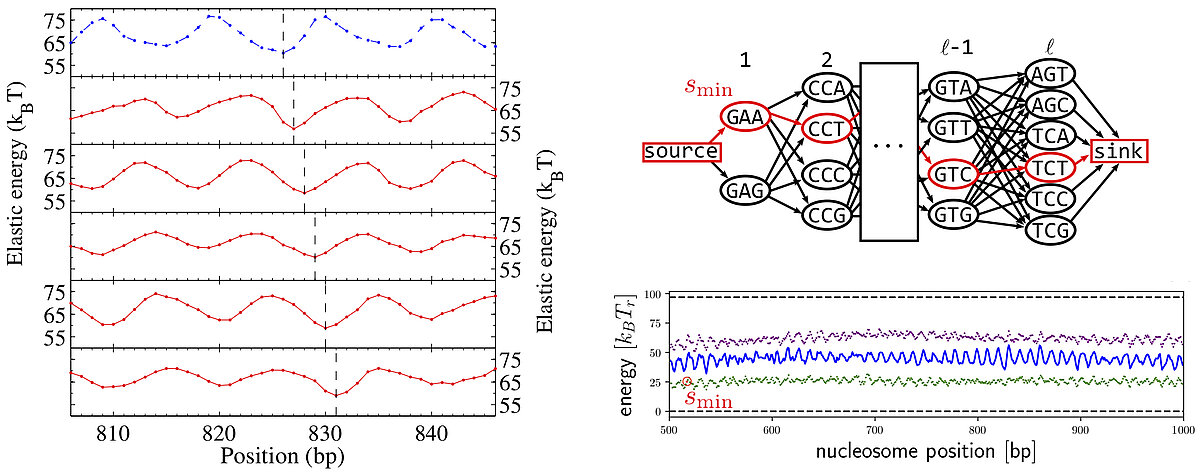
The shape and stiffness of the DNA double helix depends on the underlying sequence of base pairs. This allows to put mechanical cues on genomes that guide the folding of DNA molecules in cells. Specifically, DNA is wrapped around protein cylinders and the mechanical cues can direct the positions of the resulting complexes, called nucleosomes. Using a coarse-grained nucleosome model we were able to recover the well-known nucleosome sequence preferences (Eslami-Mossallam 2016). As our model is purely mechanical we can conclude that these sequence preference are mainly mechanical in nature.
However, DNA carries genetic information, the genes that encode for proteins. Does this mean that mechanical cues cannot be written on top of genes? We demonstrated that the degeneracy of the genetic code (64 codons encode for only 20 amino acids) allows to put cues freely on top of genes (Eslami-Mossallam 2016). This is an example of multiplexing, as we know it from daily life technology, e.g. radio channels on the same wire. Speaking of radio channels: this work has been featured at BBC World Service.