DNA is tightly packaged in each of our cells, wrapped around protein cores in the nucleus known as nucleosomes. A group of proteins powered by ATP play a huge role in changing the architecture of our DNA, by targeting nucleosomes to allow gene expression. “Chromatin remodelers are molecular motors that act on nucleosomes, moving them along DNA or assembling and disassembling them. They perform essential regulatory functions, such as controlling gene expression and DNA repair” according to Sophie Klempahn, a PhD student in the Helmut Schießel Group at the Cluster of Excellence, Physics of Life (PoL). Despite their invaluable regulatory function in cells, chromatin remodelers have received very little attention in the biophysics community. Instead, research has largely focused on nucleosomes and chromatin itself, and literature regarding the physics of chromatin remodelers modulating the genomic landscape are sparse.
Two publications were released in June of this year with Klempahn as lead author, highlighting the importance of nucleosome remodelers, in a bid to expand interest amongst biophysicists. First, a review published in Biophysical Reviews, discusses the key properties of nucleosome remodelers obtained through theoretical and experimental means. This includes the size of remodelers, their domain composition, speed of remodeling, behaviour, and the forces they exert. Recruitment of remodelers to nucleosomes has been captured by kinetic proofreading scenarios, where the combined binding of motor domains to the two gyres (regions within a nucleosome) of DNA along with the correct histone tail modification determines binding. One interesting avenue still to be explored is the role of DNA itself in the remodeling process, which is currently absent from the kinetic proofreading model. A broader understanding of remodeling mechanisms will be invaluable for progress in the field. The role of remodelers cannot be overstated, as Klempahn points out: “deregulation of these motors can lead to cell death and contribute to various diseases”.
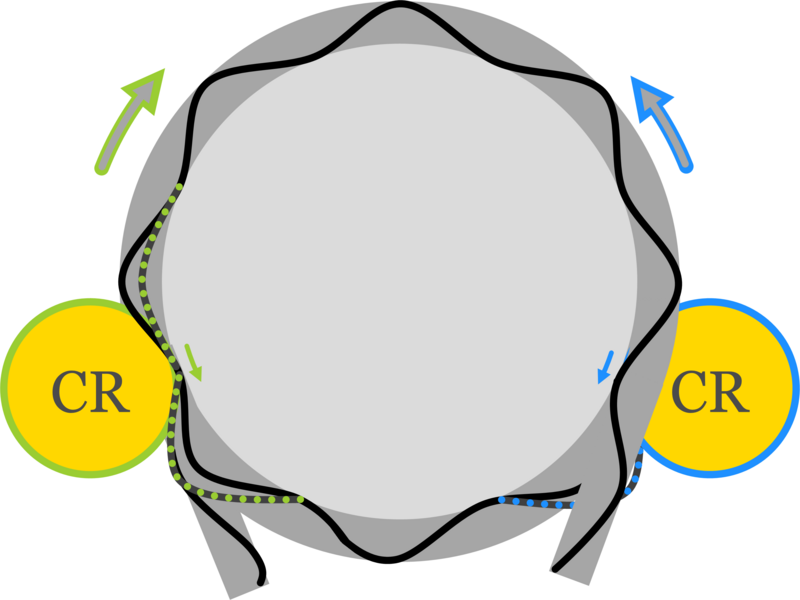
In the second publication, Klempahn and colleagues proposes a model to explore the competition between active and passive mechanisms involved in the wrapping of DNA into nucleosomes. Published in Physical Review Research, the authors build on structural insights into remodeler function and coarse-grained simulations developed from experimental data, presenting a statistical physics-based model. Authors first built a one-dimensional model mapping the 3D structure of a remodeler acting on a nucleosome, which could then be used to explain the mechanochemical cycle of nucleosome repositioning. This was aligned with previous experimental work, to confirm some remodelers operated in this manner. One key finding of the model is that chromatin remodelers push nucleosomes to specific regions of the DNA, and these actions are independent of the intrinsic preferences of nucleosomes. The authors speculate a potential “genomic code for nucleosome remodeling” where sequences may have evolved to create attractors and repellers, for remodeler-propelled nucleosomes. The work outlined here by Klempahn and colleagues is essential for our understanding of nucleosome and remodeler dynamics, and in a broader sense, how DNA is processed. Expanding our understanding of chromatin remodelers “is key to unlocking innovative treatments and insights into fundamental cellular processes”, Klempahn concludes.
Congratulations to Klempahn and colleagues on the publications! We are looking forward to more discoveries in the future.
Publications
Molecular motor in a box: Attractors, repellers, and ratchets in chromatin. Sophie Klempahn, Ralf Blossey, and Helmut Schiessel. Physical Review Research, (2024). DOI: 10.1103/PhysRevResearch.6.023236
Chromatin remodelers: a concise introduction for biophysicists. Sophie Klempahn, Helmut Schiessel & Ralf Blossey. Biophysical Reviews, (2024). DOI: 10.1007/s12551-024-01199-1